Abstract
BACKGROUND:
The pathogenesis of acute myeloid leukemia (AML) is often attributed to the presence of somatic allelic variant(s) in hematopoietic stem/progenitor cells. However, malignant clones may have heterogenous cell-surface immunophenotypes including overlap with non-malignant cells. While leukemia-associated immunophenotypes and difference from normal approaches are used for flow cytometric assessment during and after treatment, such analysis may underrepresent true leukemia disease burden. Assessments of AML measurable residual disease (MRD) using flow cytometry and molecular methods have been reported as discrepant. Single-cell RNA sequencing experiments have recently attempted to distinguish malignant cells based on gene expression and/or immunophenotypic profiles alone. We hypothesized that single-cell genotyping of mutated transcript(s) coupled with broad surface proteome and transcriptome profiling could provide an integrated multimodal method for AML characterization.
METHODS:
We adapted the previously reported "genotyping of transcriptomes" (PMID: 31270458) to identify cells carrying the NPM1 type A mutation commonly seen, and typically stable throughout the disease course, in AML. Healthy human peripheral blood mononuclear cells (PBMC) were mixed with an AML cell line carrying NPM1 type A mutation (OCI-AML3) at 7:3 ratio and labelled with 163 oligo-tagged antibodies. Single cell 3'v3 gene expression- (GEX), antibody derived tag- (ADT) and genotyping of NPM1 (GNPM) -libraries (10X Genomics) were sequenced on the NovaSeq 6000 (Illumina). Results were processed using Seurat 4.0 toolkit.
RESULTS:
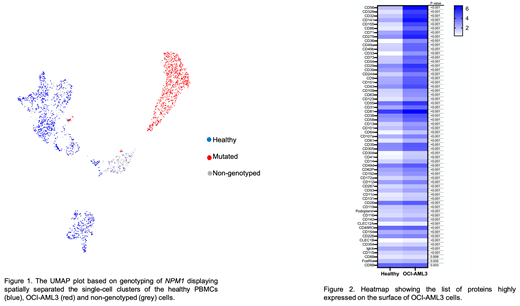
In total, 72% (n=1680) of barcoded cells could be genotyped for NPM1. Of the genotyped cells, 59% (n = 986) were not NPM1 mutated. Visualization using Uniform Manifold Approximation and Projection (UMAP) showed separation of healthy PBMCs and OCI-AML3 cells using protein data, confirmed by annotation using NPM1 genotyping (Figure 1). We found a significant positive correlation between mRNA and corresponding cell surface protein expressions in non-mutated (Pearson's coefficient, r = 0.502, p = 6.87e-11) and NPM1 mutated (r= 0.392, p= 7.5e-7) cells. Compared to non-mutated, NPM1 mutated cells showed nearly 14-fold higher NPM1 transcript levels. In addition, a total 63 proteins were highly expressed on the surface of NPM1 mutated cells (Figure 2). Among these, CD33 and CD36 showed maximum 8-fold increase in expression. Other highly expressed proteins with at least >2.5-fold change were cell adhesion molecules (including CD328, CD155, and CD56), extracellular matrix binding proteins (CD49a/b) and interleukin receptor (CD123).
CONCLUSION:
Overall, our results demonstrate proof of principle that high-throughput cell surface proteome, transcriptome and genotyping analysis can be simultaneously performed to comprehensively and confidently characterize individual AML cells. Patient-specific multiomics data with broad cell-surface proteomic screening may allow novel target identification for monitoring and/or therapeutic intervention. Ongoing work will now use this methodology to characterize a cohort of NPM1 mutated AML patient samples.
Hourigan: Sellas: Research Funding.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal